Research
This page details some of the major themes of my past research projects. For a full up to date list of papers, see publications.
AI-Derived Multimodal Cancer Imaging Biomarkers (2023-)
Recently, much of my work has focused on the use of AI to derive imaging biomarkers for cancer. The promise of precision oncology depends upon having biomarkers that predict patient outcomes and response to specific therapies to inform individualised patient decisions. Our group is interested in advancing this field using AI to integrate multiple high-dimensional data types, such as radiology imaging, pathology imaging, and genomics, which are already available from patients' standard-of-care treatment.
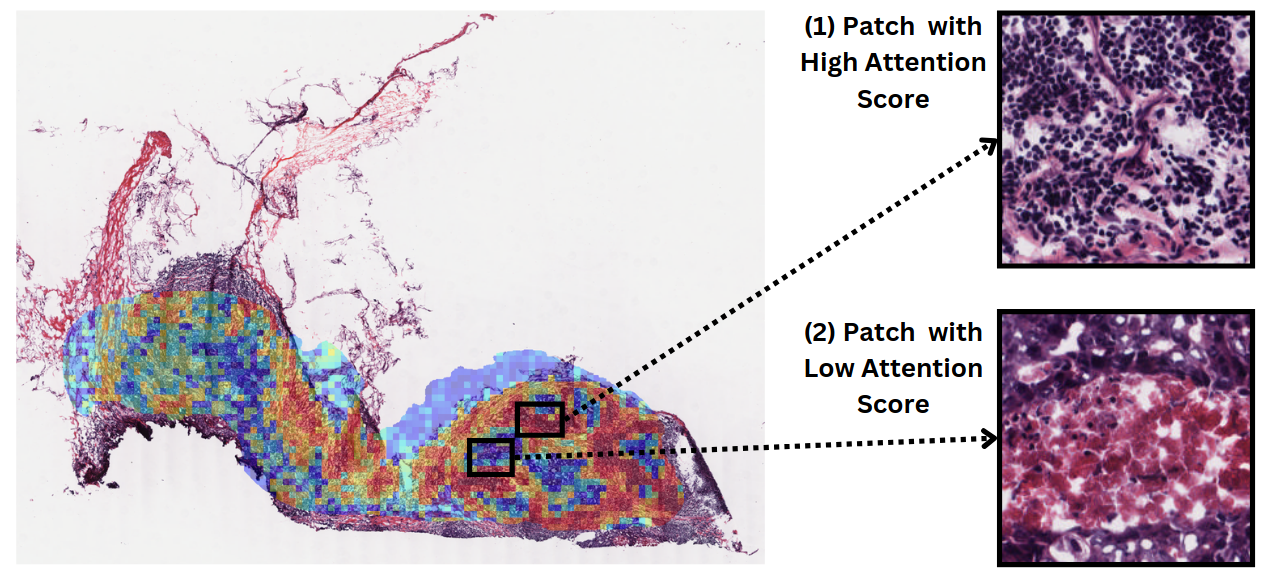
Deep learning model's attention map obtained for predicted degree of T-cell cytotoxicity from a whole slide image, demonstrating abundant lymphocytes in high-attention areas, and necrosis in low-attention regions. See the relevant paper here.
Initial publications/abstracts in this space:
- Towards Early Detection: AI-Based Five-Year Forecasting of Breast Cancer Risk Using Digital Breast Tomosynthesis Imaging
- ESPWA: A Deep Learning-Enabled Tool for Precision-Based Use of Endocrine Therapy in Resource-Limited Settings
- Deep Learning-Based Prediction of Immune Checkpoint Inhibitor Efficacy in Brain Metastases Using Brain MRI
- Enhancing Precision Oncology for Haitian Breast Cancer Patients Using Deep Learning-Enabled Histopathology Analysis
- Using Interpretable Rule-Learning Artificial Intelligence to Optimally Differentiate Adrenal Pheochromocytomas from Adenomas with CT Radiomics
- A Review of Deep Learning for Brain Tumor Analysis in MRI
- Multimodal Deep Learning-Based Prediction of Immune Checkpoint Inhibitor Efficacy in Brain Metastases
- CASCADE: Context-Aware Data-Driven AI for Streamlined Multidisciplinary Tumor Board Recommendations in Oncology
- Deep Learning-based Prediction of Breast Cancer Tumor and Immune Phenotypes from Histopathology
- From Machine Learning to Patient Outcomes: A Comprehensive Review of AI in Pancreatic Cancer
Open-Source Tooling for Standards in Medical Imaging AI (2020-)
As artificial intelligence models become part of standard clinical imaging workflows, the use of appropriate data standards to communicate AI results becomes vital to ensure that AI tools can interoperate with clinical systems. In particular, results derived using AI must be accompanied by appropriate metadata for correct use within a clinical environment. While the DICOM standard is ubiquitous throughout radiology (and present to some extent in other medical specialisms that use imaging such as pathology and ophthalmology), using DICOM format files to store files is a complex task for most machine learning scientists.
To close this gap, the highdicom Python library, which I authored and maintain along with my colleague Markus Herrmann, aims to provide tools for AI developers to both create and parse DICOM files containing image-derived information (such as segmentation masks, measurements, classifications and regions of interest) and use them directly in AI training processes. The library is used by researchers and developers all around the world and has been downloaded hundreds of thousands of times.
Beyond the library, I am interested in moving medical imaging AI workflows to be more closely aligned with clinical standards and the DICOM format in particular at all stages of the model development process. I am currently working with the National Cancer Institute's Imaging Data Commons initiative to enable the dissemination of large-scale cancer imaging datasets.
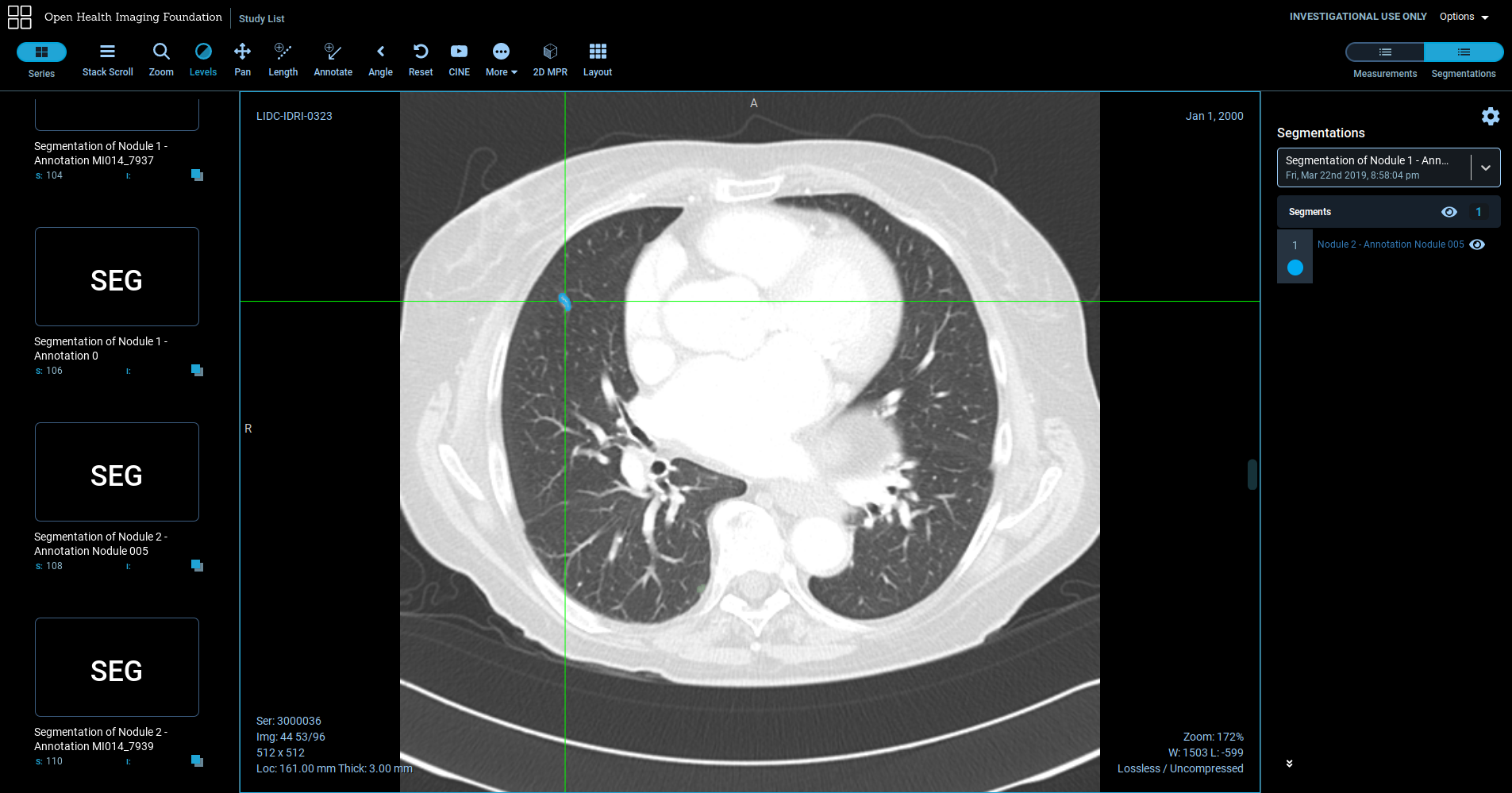
Lung nodule segmentation stored as a DICOM Segmentation object using the highdicom library and visualised using the open-source OHIF viewer.
See our paper “Highdicom: A Python Library for Standardized Encoding of Image Annotations and Machine Learning Model Outputs in Pathology and Radiology” for more details about the libary, and “NCI Imaging Data Commons: Towards Transparency, Reproducibility, and Scalability in Imaging AI” for a description of the Imaging Data Commons.
Development of Machine Learning Medical Imaging Analysis Models in a Translational Setting (2017-2022)
The majority of my time working at the Mass General Brigham Data Science Office was focused on the development of medical imaging AI models targetted for commercial translation and deployment. Some of these models are not yet publically disclosed, but others have led to scientific manuscripts. One particular early focus was on models to automate rapid image interpretation along the clinical stroke workflow, where speed of image interpretation is particularly important. We developed models for quantification of ischaemic infarct volume in diffusion MRI, detection of acute ichemic stroke in non-contrast CT, and detection of large vessel occlusions in CT angiography images. I currently have three patents pending and multiple models going through the process of review at the US Food and Drug Administration for commercialisation.
Related publications:
- Development and Clinical Application of a Deep Learning Model to Identify Acute Infarct on Magnetic Resonance Imaging
- Head CT Deep Learning Model is Highly Accurate for Early Infarct Estimation
- Enhanced Physician Performance When Using an Artificial Intelligence Model to Detect Ischemic Stroke on Computed Tomography
- (Patent application) Computed Tomography Medical Imaging Intracranial Hemorrhage Model
- (Patent application) Medical Imaging Stroke Model
- (Patent application) CTA Large Vessel Occlusion Model
Opportunistic CT Body Composition Analysis (2018-)
Body composition describes the proportions and distribution of tissues such as fat and muscle in the body. A person's body composition is an important biomarker and is related to their risk of various medical conditions such as stroke and heart attacks and the likelihood of complications during surgical procedures. Body composition can be inferred ‘opportunistically’ using segmentation of routine chest/abdomen CT scans, but this is too slow to be practical for clinicians to perform routinely. If however, we can use automated methods to extract body composition from routine scans, we can make these important biomarkers available at scale within routine care.
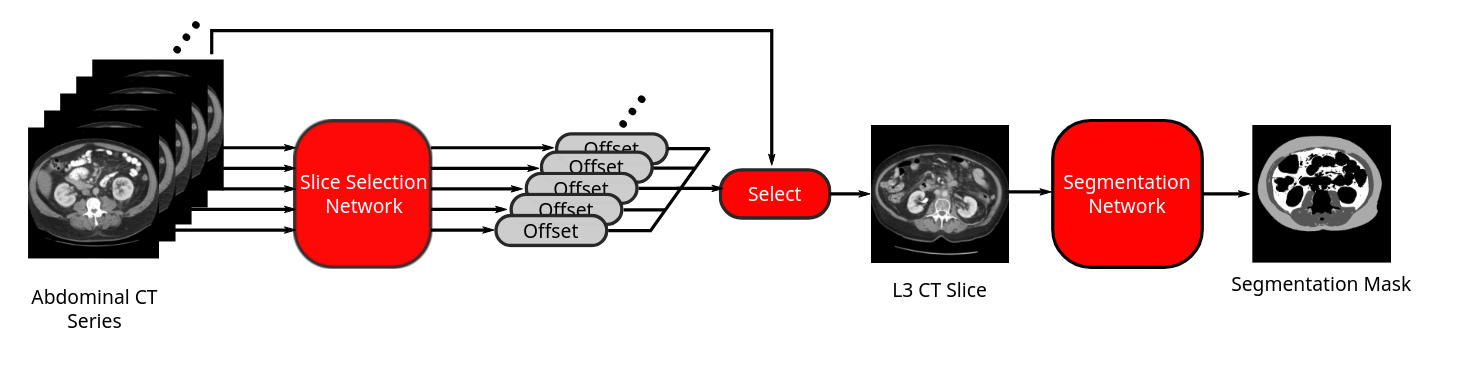
Flow diagram of our fully-automated end-to-end body composition analysis pipeline from abdominal CT. Figure: Bridge et al 2018
Working with a multi-disciplinary team at the MGB Data Science Office, Brigham and Women's Hospital, Dana Farber Cancer Institute, and Massachusetts General Hospital, I developed a fully-automated machine-learning-based method using two convolutional neural networks to first locate a suitable slice at the L3 level for analysis, and then segment muscle, subcutaneous, and visceral fat from abdominal CT scans. I then further developed this method to analyse further thoracic levels to broaden the appliability to chest CTs. The complete algorithm can run efficiently and in a fully-automated manner on large cohorts of CT to enable large scale research studies. In this way, I have since collaborated with a number of groups to establish population reference curves for body composition in adults and demonstrate the utility of body composition biomarkers for applications as diverse as oncology, surgery, cardiovascular health, and frailty.
Related publications include:
- Fully-Automated Analysis of Body Composition from CT in Cancer Patients Using Convolutional Neural Networks
- A Fully Automated Deep Learning Pipeline for Multi-Vertebral Level Quantification and Characterization of Muscle and Adipose Tissue on Chest Computed Tomography
- Population-Scale CT-Based Body Composition Analysis Of a Large Outpatient Population Using Deep Learning To Derive Age, Sex, and Race-Specific Reference Curves
- Multilevel Body Composition Analysis on Chest Computed Tomography Predicts Hospital Length of Stay and Complications Following Lobectomy for Lung Cancer: A Multicenter Study
- Computed Tomography-Based Body Composition Profile as a Screening Tool for Geriatric Frailty Detection
- Evaluating Frailty, Mortality, and Complications Associated with Metastatic Spine Tumor Surgery Using Machine Learning-Derived Body Composition Analysis
- Utility of Normalized Body Composition Areas, Derived From Outpatient Abdominal CT Using a Fully Automated Deep Learning Method, for Predicting Subsequent Cardiovascular Events
- Role of Machine Learning-Based CT Body Composition in Risk Prediction and Prognostication: Current State and Future Directions
- Body Composition and Lung Cancer-Associated Cachexia in TRACERx
Ultrasound Video Analysis (2013-2017)
The main focus of my doctoral research was the automated analysis of ultrasound videos, with a particular focus on fetal heart scanning. Previous work has been successful in automatically detecting objects of interest in still ultrasound images, which can be useful for post-acquisition measurement on typically carefully-selected images. My aim was to extend this to full video analysis, working towards creating software that can, to some degree, understand what is happening in the scan’s video stream as the scan is under way. This has the potential to provide software tools to assist sonographers acquiring scans, and thereby improve the effectiveness of ultrasound as a screening process.
I advocate the use of Bayesian filtering techniques as a natural framework to use in this context. The key advantages of this group of probabilistic techniques are a) they naturally deal with the uncertainty that is inherent in the interpretation of the video, and b) allow an engineer to build in prior knowledge of how ultrasound videos typically evolve in order to combine information from multiple frames into a consistent hypothesis about the entire video. This is crucial in the setting of ultrasound video analysis, where the single video frames can be quite uninformative due to the indistinct nature of the imagery and the presence of imaging artefacts, but the spatial and temporal relationships between the variables of interest are very strong. Similar techniques have proved popular in situations such as 3D human pose tracking from video footage, and localisation of mobile robots in uncertain environments (the so-called ‘SLAM’ problem).
In my journal paper “Automated Annotation and Quantitative Description of Ultrasound Videos of the Fetal Heart”, I present and evaluate a Bayesian filtering framework for analysing fetal heart videos. The algorithm uses stochastic methods (particle filters) to represent and solve the filtering problem. The video above shows some examples of the output of this algorithm (top) compared to the manual annotations (bottom). Comparing the “no filtering” and “with filtering” examples shows how important the filtering framework is for achieving good results.
In more recent work (“Localizing Cardiac Structures in Fetal Heart Ultrasound Video”), I extended this framework to additionally track diagnostically important anatomical structures such as valves and vessels, as shown in the video below.
Along with colleagues, I have begun to look at deep learning as an alternative method for ultrasound video analysis.
This work was supervised by Alison Noble at the Institute of Biomedical Engineering, University of Oxford, and was performed in collaboration with Christos Ioannou (John Radcliffe Hospital, Oxford).
Analysis of Fetal Heart Ultrasound Videos (2013-2017)
For my work on ultrasound video analysis, I have focused on videos of the fetal heart. Ultrasound screening is the standard clinical method for antenatal detection of congenital heart defects, such as septal defects, vessel coarctation, tetralogy of Fallot, arrhythmias, and others. Unfortunately, screening for these defects is a highly skilled task as it involves checking a number of different anatomical structures in multiple viewing planes, and, as such, detection rates vary.
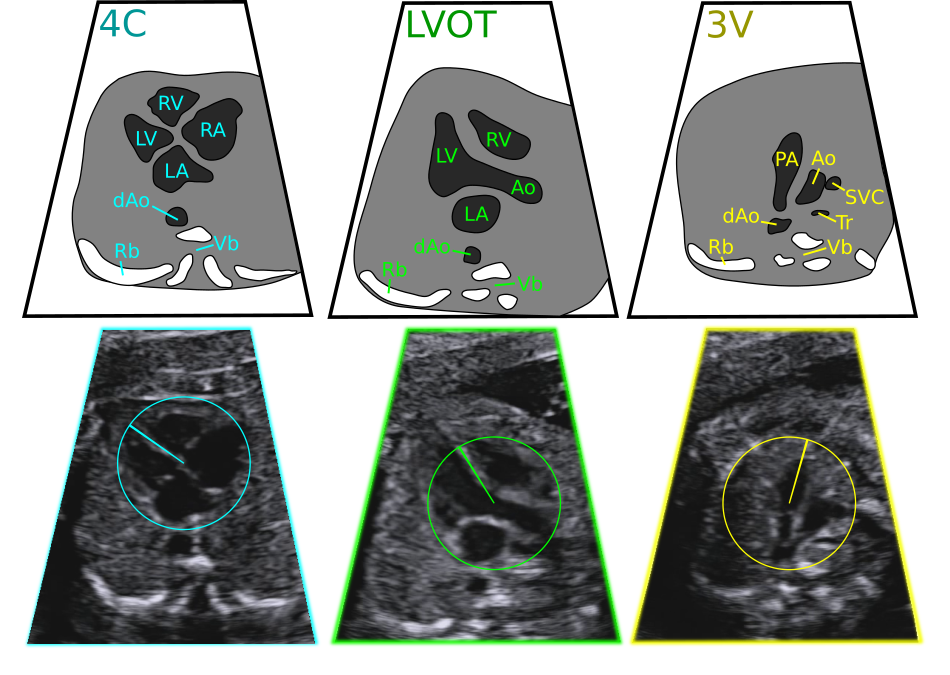
Three fetal heart viewing planes: the four chamber view (4C), left ventricular outflow tract view (LVOT), and the three vessels view (3V). Key: LV/RV left/right ventricle, LA/RA left/right atrium, (d)Ao (descending) aorta, PA pulmonary artery, SVC superior vena cava, Tr trachea, Rb rib, Vb vertebra.
Using Bayesian filtering, I have developed software that tracks the fetal heart, including its position, orientation, viewing plane, and the position in the cardiac cycle. I hope that this could aid sonographers during the acquisition process and provide a basis for more specific automated diagnostic screening processes.
Rotation Invariant Features in Ultrasound Image Analysis (2013-2017)
Many traditional computer vision algorithms are designed to detect objects that occur in images in a small range of orientations. However, in a number of medical imaging applications, this approach is not appropriate. This is particularly true in fetal ultrasound imaging, where the orientation of the fetus relative to the probe varies within and between scanning sessions.
Instead of simply applying traditional techniques to multiple rotated versions of the image, I have investigated more principled approaches to rotation invariant detection that build rotation invariance into the features used by machine learning algorithms.
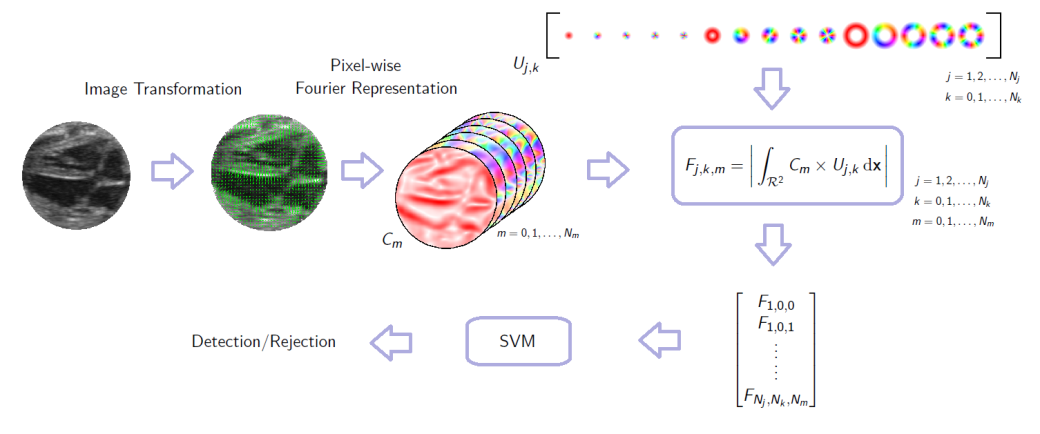
A framework for rotation-invariant detection using image gradient. First a continuous gradient orientation histogram is formed and represented by a small number of Fourier series coefficients. These coefficient images are then convolved with a set of complex-valued basis functions. The phase of the resulting complex number is invariant to the rotation of the underlying patch.
I use complex-valued filters to create descriptive feature sets for image patches that are analytically invariant to the rotation of the patch (this follows the work of Liu et al.). This forms a description of the patch that may be used for classification/detection and other machine learning tasks. This is described in detail in “Object Localisation In Ultrasound Images Using Invariant Features”.
The Monogenic Signal for Medical Image Analysis (2013-2015)
The monogenic signal is an image representation with a number of attractive properties for tasks such as edge and feature detection. It has been found to be particularly useful for analysing ultrasound images, because it copes well with the variable contrast, speckle artefacts and indistinct features in the images. It also forms the basis for calculating a number of other quantities such as local phase, local amplitude/energy, local orientation, feature symmetry, feature asymmetry and phase congruency.
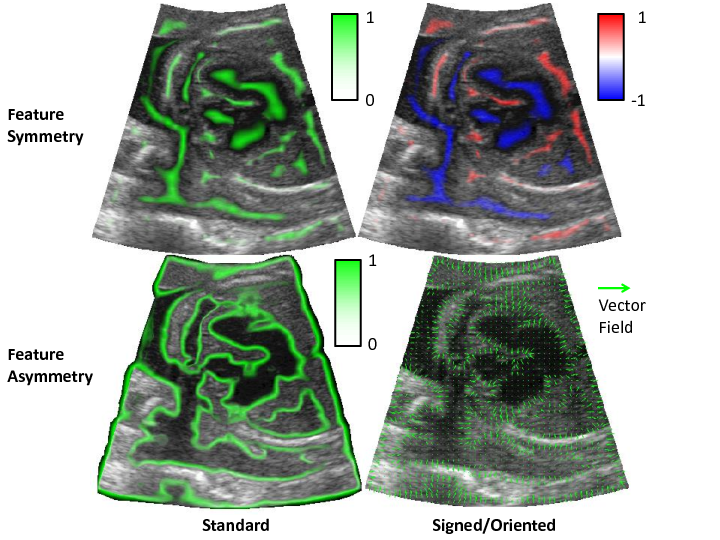
Feature symmetry (top) and asymmetry (bottom) may be calculated from the monogenic signal and are useful for detecting ‘blob’ structures and boundaries respectively in images. © IEEE 2015
In trying to understand many of the concepts surrounding the monogenic signal I found that the current literature does not offer any introductory text, and that this makes the area somewhat impenetrable. I therefore wrote a tutorial-style “Introduction to the Monogenic Signal” document to help explain the concepts to others in my lab.
In my paper “Object Localisation In Ultrasound Images Using Invariant Features”, I demonstrated an effective detection framework for object in ultrasound images that makes use of the monogenic signal to increase invariance to speckle artefacts and contrast changes.
Registration of the Human Femur (2012-2013)
The thickness and distribution of femoral cortical bone is thought to be an important factor determining a person’s susceptibility to hip fracture. Cortical thickness can be estimated from a CT scan, giving a surface representation of a subject’s femur with associated cortical thickness values.
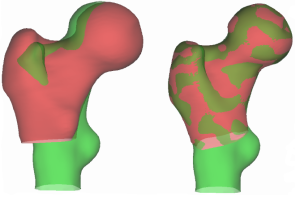
Example of a registration failure, where the lesser trochanters on the two surfaces are not aligned.
However, in order to conduct large-scale quantitative medical studies, it is necessary to first find the correspondence between points lying on the different surfaces. This requires a reliable, automated registration procedure. In my fourth-year undergraduate research project, I investigated and implemented a number of improvements to an existing registration procedure.
One contribution was testing a ‘locally affine’ transformation, which I found to reduce unnecessary warping during the registration procedure due to its local nature.
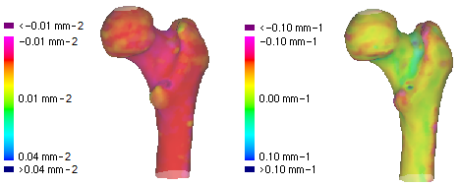
Gaussian (left) and mean (right) curvature quantify local surface shape.
A second important contribution was incorporating surface differential geometry into the cost function of the point matching. This ensures that the aligned points on the two surfaces have similar local geometry.
The outcome of this work was improvements to the ‘wxRegSurf’ software, freely available at the project website along with more information, pretty pictures and lists of related publications. You can also read my [Project Report] or [Presentation].
This work was conducted under the supervision of Andrew Gee at Cambridge University Engineering Department.